58 salps representing the species Salpa thompsoni, Pegea confoederata, Thalia democratica, Soestia zonaria, Thetys vagina, Salpa fusiformis, and Ihlea magalhanica, including both solitary and aggregate stages of the first four and a distribution of sizes for the first two, were collected during the "SalpPOOP" cruise on R/V Tangaroa. Salps were collected via twice daily oblique Bongo tows down to 200 meters as well as daily ring net surface tows with a 30-liter non-filtering cod-end. Once onboard, salps from ring net and Bongo tows were identified to the species level, sorted by life stage (i.e., solitary or aggregate), measured, and sexed (Foxton et al. 1966; Lüskow et al. 2020). Triplicate representative samples for each salp species from a total of 10 of these casts were preserved in 5% formalin < 30 minutes after collection the first time each species was encountered.
Once ashore, SEM samples were prepared from each preserved organism by excising guts under a HEPA-filter equipped laminar flow exhaust hood using acid-cleaned plastic dissection equipment to minimize contamination. Guts were then placed in either 15 or 50 mL plastic Falcon tubes with a small volume of brine, lacerated, and then vortex mixed for two minutes to release gut contents into solution while minimizing damage to the more fragile phytoplankton (Jung et al. 2010; von Harbou et al. 2011; Ahmad-Ishak 2017). An aliquot of this solution was then filtered onto a 0.2 μm Nuclepore filter, followed by six rinses of decreasing salinity in 5 ppt increments for a minimum of 5 minutes each with the final MilliQ water rinse performed twice. This was immediately followed by a dehydration series of increasing ratios of Ethanol:MilliQ to purge water from the sample, with the final 100% anhydrous ethanol step again performed twice. Finally, a substitution series of increasing ratios of the chemical drying agent hexamethyldisilazane (HMDS):anhydrous ethanol was conducted with each step lasting a minimum of 10 minutes, with the final HMDS step being allowed to air dry. Each step was conducted under either a light vacuum or gravity filtration depending on material concentration to minimize loss between treatments. The dried filter was then affixed to an aluminum SEM stub using carbon conductive adhesive tabs and further grounded with a thin piece of carbon conductive tape touching the edge of the filter and the bottom of the stub. Samples were then sputter coated with 10 nm iridium and visualized using an FEI Nova 400 NanoSEM set to an accelerating voltage of 10 kV. Twenty random regions of each filter were imaged at 3 different magnifications: ~500x, ~2,500x, and ~12,000x to target microplankton (20-200 µm), nanoplankton (2-20 µm), and picoplankton (<2 µm), respectively.
Particles in SEM images were manually outlined using ImageJ (v. 1.52a or 1.53c) to extract the maximum feret length (Feret.um), minimum feret length, and area (Area.um2). These measurements were used to estimate equivalent spherical diameter (ESD.um), biovolume (BV), and carbon biomass assuming a prolate spheroid. To avoid overestimating the size of irregularly shaped particles, we calculated width (Width.um) for a prolate spheroid of measured area and length such that:
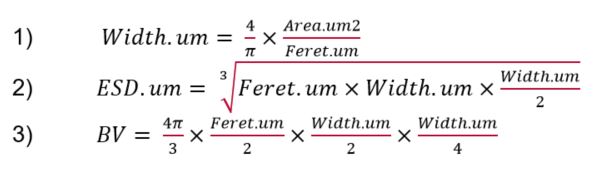
(Note: The equations in this image are also attached as Supplemental File "SalpGut_Particles_Equations.JPG".)
Note that because we estimated the three-dimensional size of particles using a two-dimensional image, we determined the height of each particle from its width. To account for the ~50% flattening of height caused by filtration (Taylor et al. 2011), we furthermore assumed height to be equivalent to half of the width. This correction was only applied for soft-bodied groups such as picoplankton and most nanoplankton, while for incompressible particles such as diatoms and dinoflagellates no correction was applied.
Since sufficient structural detail could not be observed to definitively identify very small particles, spherical particles within the size range of ~0.4-1.5 µm and resembling control images from lab cultures of Prochlorococcus sp. and Synechococcus sp. are instead referred to as bacteria-like particles (PartType "BLP"). Other PartTypes include: “WS” for white spheres assumed to be partially digested nanoflagellates; “Unknown” for nondetrital particles for which a classification could not be provided; "Alex" for Alexandrium spp.-like dinoflagellates; "ProB" for Prorocentrum minimum; "ProD" for Prorocentrum dentratum; "UnkDino" for unknown dinoflagellates; "Cyst" for particles resembling the resting stage cysts of diatoms; "Oxy" for Oxytoxum spp.; "Cerat" for Ceratium spp.; "CeratSpike" for only a portion of a Ceratium horn; "Tin" for tintinnids; "Pennate" for unknown pennate diatoms; "Dicty" for Dictyocha speculum; "UnkCocc" for particles similar to coccolithophores but with distinct holes at regular intervals (possibly just digestion artefacts); "Centric" for unknown centric diatoms; "SmallPen" for smaller, possibly nitzschiaform diatoms that smaller and fatter about the center than "Pennate"; "Diplo" for Diplopelta spp.; "Cocco" for unknown coccolithophores; "Frag" for Fragilariopsis spp.; "Radio" for unknown polycystine radiolarians; "Pnut" for unknown pennate diatoms with shape reminiscent of a peanut; "Pyro" for Pyrodinium spp.; "RaphDia” for unknown pennate diatom with distinct raphid ridge; "Dinoph" for Dinophysis spp.; "Ciliate" for unknown ciliates. Each class may also be proceeded by a "B", connotating the particle was considered broken or missing more than ¾ its true size. "CeratSpike" was considered broken as well.
The biomass of formalin-preserved ciliates was estimated as 0.14 pg C µm⁻³ (Putt and Stoecker 1989) while rhizarians were 0.001 pg C mm⁻³ (Stukel et al. 2018). The biomass of diatoms was estimated allometrically as 0.288*BV^0.811 while other protists and unidentified particles were estimated using 0.216*BV^0.939 (Menden-Deuer and Lessard 2000). Because we could not differentiate between types of prokaryotes in the SEM images, we calculated a single average biomass conversion for all bacteria-like particles in the salp guts using published allometric relationships weighted by the ratio of each of the key bacterial groups to each other in the water column from flow cytometry data for a given cycle.

 ©2025 Biological and Chemical Oceanography Data Management Office.
©2025 Biological and Chemical Oceanography Data Management Office.
